Research
Evolutionary dynamics in the human gut microbiome
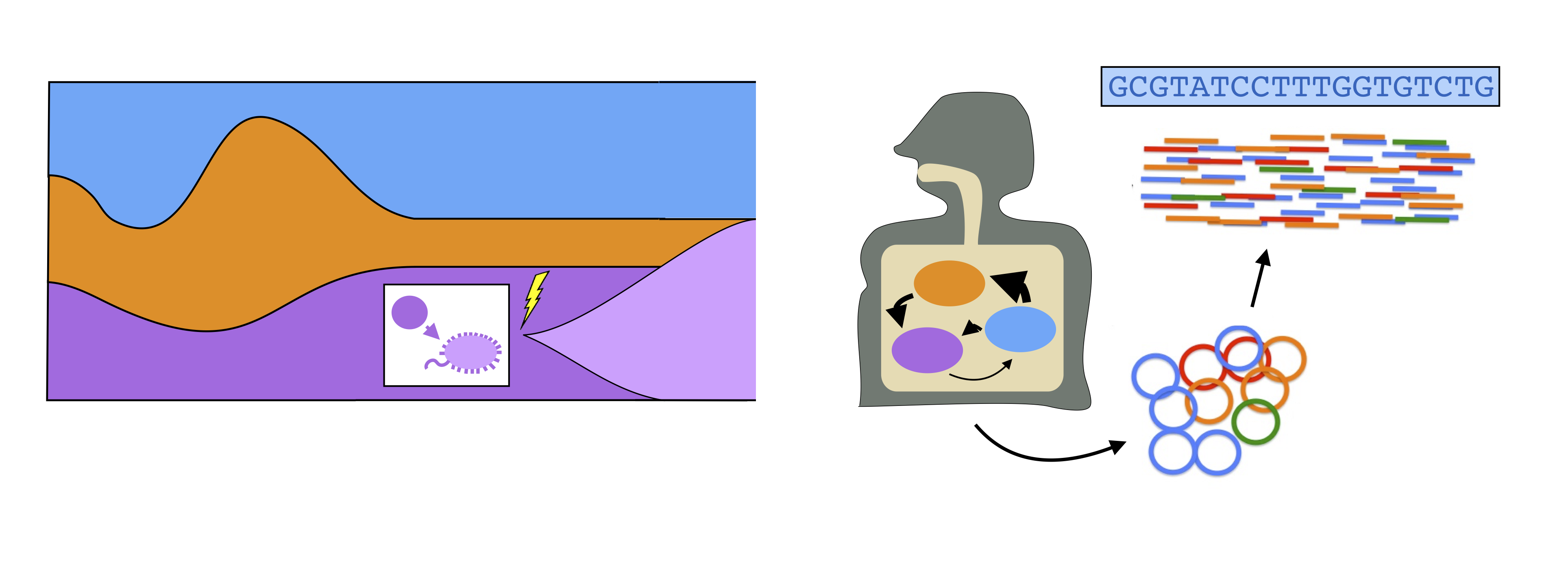
The human gut harbors a diverse microbial community, which performs a number of critical health-related functions. High rates of cell turnover endow these organisms with an enormous potential for rapid evolutionary change: more than a billion new mutations are thought to arise within a single host community every day. Despite the potential importance of these effects, we currently know very little about the evolutionary dynamics that occur within individual species in this community. This robs us of one of our best empirical opportunities to understand how evolution operates in a complex community setting.
To bridge this gap, we are developing computational approaches for measuring evolutionary changes within species in complex metagenomic datasets, and for interpreting these patterns using simple null models from population genetics. Our focus is on identifying the relevant timescales and population genetic “rules” that govern the evolution of gut bacteria in their native habitat, and in leveraging these empirical constraints to guide further theoretical and experimental efforts. Current topics of interest include:
- How does the gut microbiome respond to antibiotic perturbations at the genetic level?
- How relevant are different population genetic forces like natural selection, genetic drift, and horizontal gene transfer within hosts? How do these within-host processes interact with longer-term variation across hosts?
- How does within-species evolution impact the broader ecological strutcure of the community? (and vice versa?)
Eco-evolutionary dynamics and genetic diversity in resource competition models
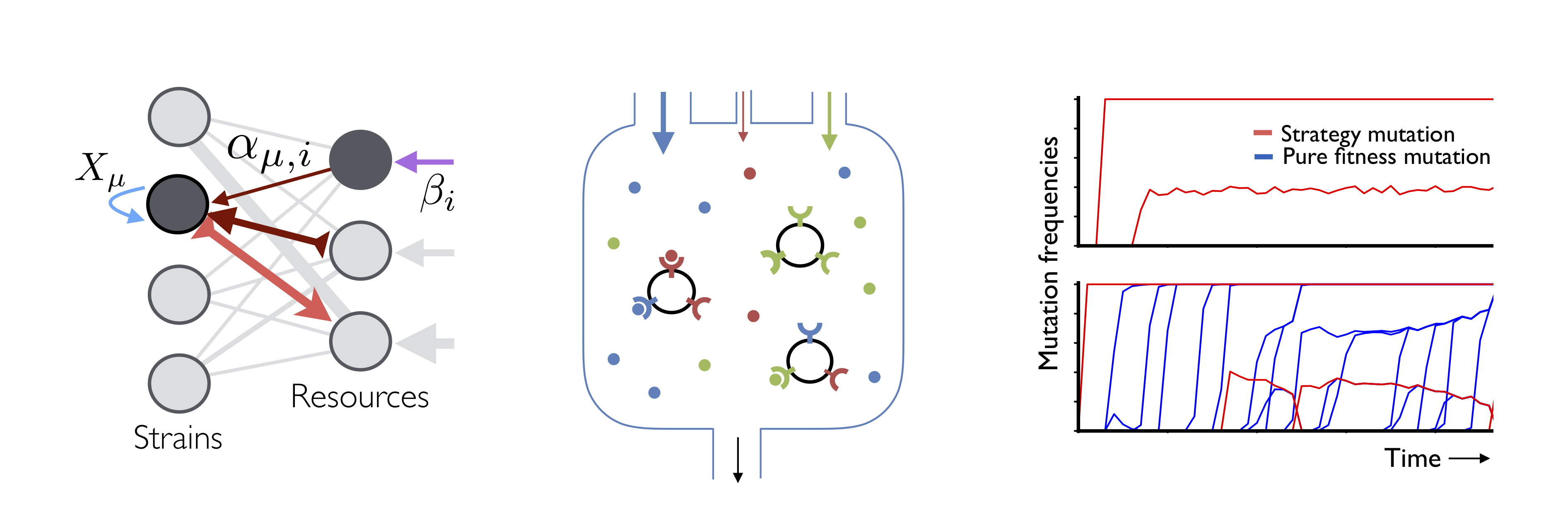
A major barrier to understanding evolution in microbial communities is that we have few theoretical tools to describe situations where ecology and evolution interact. Traditional ecological theory describes the competition between strains, but the strains themselves are rarely allowed to evolve over time. Conversely, population genetic theory describes the evolution of DNA sequences, but the relevant “populations” must be fixed externally. The interactions between these processes have traditionally been explored in computer simulations, but the vast number of parameters makes it difficult to extrapolate from these individual examples, or to understand how they map on to experiments. To make further progress, it will be critical to develop simpler effective models of evolving ecosystems whose limiting behavior can be characterized analytically.
In pursuit of this goal, we are developing new theory for predicting the coupled ecological and evolutionary dynamics that emerge in simple models of resource competition. We are particularly interested in the non-equilibrium dynamics that are encoded in the statistics of sampled DNA sequences, and in our ability to extrapolate to the high-dimensional ecosystems that are often encountered in nature. Current topics of interest include:
- What are the first evolutionary steps taken by a randomly assembled community? What does the ecological and genealogical structure of a community look like after an infinite amount of evolution?
- How does evolution within a local community compete with migration between communities in a larger metapopulation?
- What are the effects of clonal interference (see below), which is known to play an important role in large microbial populations?
- How can we connect these simple toy models with the complex growth dynamics of real microbial cells? Which aspects of this complexity matter? How can we measure the relevant effective parameters experimentally?
Fundamental questions in population genetics: selection, linkage, and recombination
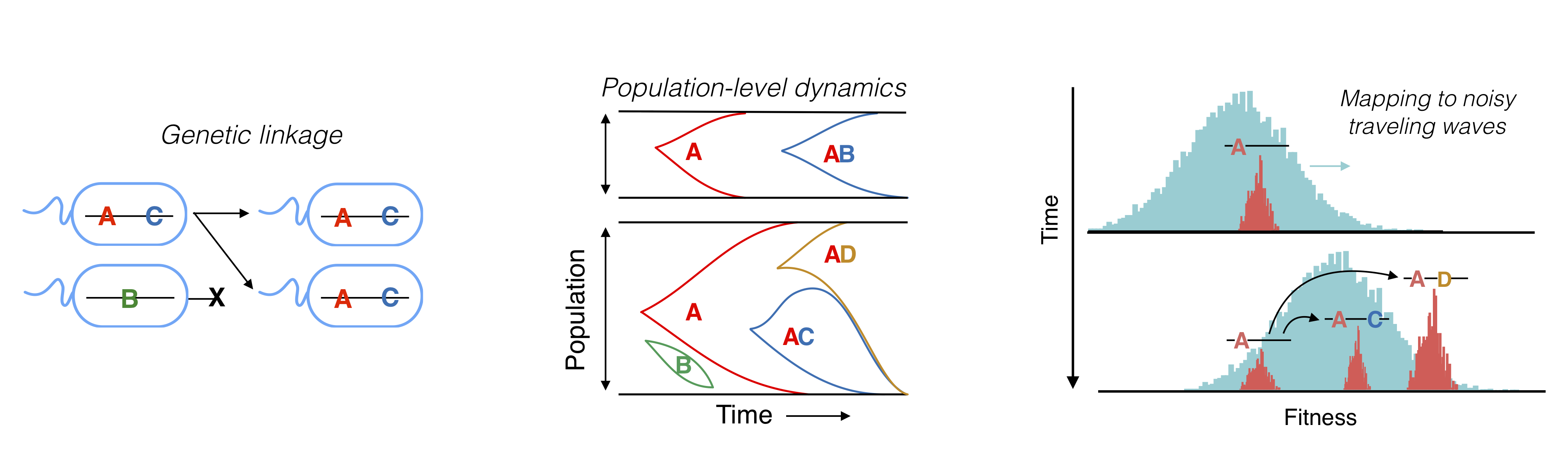
In large populations, natural selection can’t act on mutations individually, but only on combinations of mutations that happen to be inherited together on the same DNA molecule. This simple fact (known as genetic linkage) creates complex correlations along the genome that are still only partially understood theoretically. This is a major barrier in our ability to interpret data from laboratory evolution experiments and many natural populations. We work on a variety of theoretical problems to understand evolutionary dynamics and DNA sequence variability in the presence of genetic linkage. Current topics include:
- Evolution of mutation rates and other “evolvability” modifiers
- How do genetically diverse populations navigate rugged fitness landscapes?
- How does the chunk-like nature of bacterial recombination influence expected patterns of genetic diversity?
- Pairwise and higher-order correlations between mutations (“linkage disequilibrium”)